Abstract
Comparative effectiveness research (CER) in genomic medicine (GM) measures the clinical utility of using genomic information to guide clinical care in comparison to appropriate alternatives. We summarized findings of high-quality systematic reviews that compared the analytic and clinical validity and clinical utility of GM tests. We focused on clinical utility findings to summarize CER-derived evidence about GM and identify evidence gaps and future research needs. We abstracted key elements of study design, GM interventions, results, and study quality ratings from 21 systematic reviews published in 2010 through 2015. More than half (N = 13) of the reviews were of cancer-related tests. All reviews identified potentially important clinical applications of the GM interventions, but most had significant methodological weaknesses that largely precluded any conclusions about clinical utility. Twelve reviews discussed the importance of patient-centered outcomes, although few described evidence about the impact of genomic medicine on these outcomes. In summary, we found a very limited body of evidence about the effect of using genomic tests on health outcomes and many evidence gaps for CER to address.
Genet Med advance online publication 13 April 2017
Similar content being viewed by others
Main
Genomic medicine (GM) uses information about a person’s genome to improve his or her health. Growing interest in GM coincides with heightened recognition of the need for better-quality evidence to support informed decisions. Therefore, the widespread adoption of GM into patient care will require high-quality evidence that it improves patient outcomes when compared with conventional care. A third trend, which may influence the research in genomic medicine, is the inclusion of study outcomes that patients say are of greatest concern to them.
This focus on patient-relevant outcomes is the hallmark of comparative effectiveness research (CER). The purpose of CER is to improve the evidence base for making decisions that are relevant to patients and other stakeholders. CER encompasses the synthesis of existing evidence and the generation of new evidence that compares alternative approaches to the prevention, diagnosis, or treatment of a health condition. In the context of genetic tests, CER is applicable both in the analysis of individual studies and when conducting systematic reviews of a body of evidence. Specifically, after establishing the analytic validity (reliability in clinical laboratory practice) and clinical validity (diagnostic or prognostic accuracy) of a test, CER is the approach for determining how the use of the test impacts health outcomes compared with an appropriate alternative (no testing or a comparison test).
The purpose of this review is to identify opportunities for CER to contribute to making the application of GM to patient care more evidence-based and more patient-centered. Previous studies have proposed a conceptual framework or focused on specific topics such as cancer tumor profiling.1,2,3,4,5,6,7,8 We summarize the findings of systematic reviews of CER of specific GM interventions (“structured review”) and use expert assessment of these findings to help identify evidence gaps (“landscape analysis”). Structured literature reviews map the literature landscape by identifying what evidence is available and assessing the findings with respect to gaps in the evidence,9 whereas landscape analysis provides an overview of a topic by combining a structured literature review with expert input.10
Methods
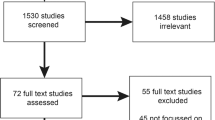
We abstracted and then summarized information from each included systematic review ( Figure 1 ). A technical working group (TWG) with GM expertise (the members are listed in the Supplementary Materials online) reviewed this information and suggested ways to portray the current landscape of GM. Our research questions were as follows:
-
1. What is the evidence from systematic reviews of CER of GM?
-
a. What tests, testing indications, comparators, and outcomes have been studied?
-
b. What is the impact of GM on patient outcomes (clinical utility)?
-
c. Did the systematic reviews use standard methods to evaluate the quality of the studies and what did they conclude?
-
d. What did the reviews say about the potential clinical role of the GM interventions?
-
e. Did any studies use patient-reported outcomes (e.g., impact on activities of daily living)?
-
f. Did the reviews identify gaps in the evidence about patient outcomes? What CER might address those gaps?
-
2. When taking into account both the evidence from the systematic reviews and expert assessments, what is the current state of CER of GM and its future?
-
Study design. We conducted a structured literature review by abstracting information from each included review and then summarizing the results (Table 1; Supplementary Materials online). The reviews, along with interviews and assessments by the Technical Working Group, were used to develop the landscape analysis.
Structured review
We systematically identified, selected, and appraised systematic reviews conducted by technology-assessment groups (TAGs), which we defined as organizations that assess health technologies to support clinical practice guidelines or insurance coverage decisions. We limited ourselves to systematic reviews because they have several desirable characteristics. Most importantly for assessing the quality of the body of evidence (our key objective), they use widely accepted quality-assessment instruments that incorporate measures of the components of study quality. They also choose topics that are important to the public at large. For example, the Blue Cross Blue Shield Association Technology Evaluation Center (BCBSA TEC; now called BCBSA Evidence Street) performs systematic reviews for its independent Medical Advisory Panel, and other organizations11 use its reports, and the Agency for Health Care Research and Quality (AHRQ) uses public input to inform the choice of topics for systematic reviews. We focused on systematic reviews because they are comprehensive by design, bias-free in their conduct, and collectively summarize a vast body of individual studies.
We describe our approach to identifying systematic reviews in Figure 2 and the Supplementary Materials online. We first developed a list of potential TAGs based on our review of the literature and confirmed this list with the TWG. Using inclusion criteria, we narrowed this to 13 key TAGs (Supplementary Materials online). We searched TAG websites using the key words “personalized medicine,” “precision medicine,” or “genomic testing” and by reviewing report titles. We included a systematic review if it summarized multiple studies of CER that addressed GM tests (as of December 2015). We limited our search to reviews from the past 5 years, which is an empirically derived interval after which a systematic review is considered outdated.12
We found relevant systematic reviews by BCBSA TEC (9 reviews), AHRQ (10 reviews), and the Cochrane Collaboration (2 reviews). Only two CER reviews were older than 5 years. All 21 performed at least a Medline search, and five performed a gray literature search. All had prespecified inclusion and exclusion criteria. In all the AHRQ and Cochrane reviews, two individuals independently extracted data from individual studies. Fourteen systematic reviews described the standards used to rate the risk of bias (Supplementary Materials online).
Data abstraction
We abstracted variables chosen to reflect study objectives and approaches used to prioritize topics for CER13 and TWG input (Supplementary Materials online). We coded reviews using the test(s) included in the review as the unit of analysis, not each individual study within the review. We categorized the reviews according to clinical testing indications (e.g., cancer tumor profiling tests). Four authors (K.P., P.D., H.S., and M.D.) summarized each review. One author (H.S.) assessed the quality of the methods used in individual studies within reviews by describing the instruments used to assess quality and recording the overall quality ratings of the evidence ( Table 1 ). We assessed whether our conclusions were likely to have been different if we had reviewed individual studies versus TAGs using two approaches. First, we determined whether the TWG reviewed all key conclusions and noted any outdated findings. Second, we compared conclusions from an included TAG review of sequencing tests for prenatal screening to recent systematic reviews not included in our study because they were not TAG reviews14,15,16 as well as to key individual studies.17,18 We found that these conclusions were similar and would not have substantively changed the findings of our study.
Synthesis of the body of evidence
We first obtained background information by conducting semistructured interviews with each TWG member and four other experts, including one patient advocate (Supplementary Materials online). Later, we asked TWG members to identify related GM tests that had not been subjected to CER and important changes in the evidence since the last study included in the 21 reviews. TWG members also commented on the CER questions. Responses reflected the individual TWG member’s opinion, not a formal process of priority setting.
Results
Table 1 lists the 21 reviews along with their stated objectives and their evidence quality ratings (details in Supplementary Materials online). Every review found methodological shortcomings and little or no evidence about the impact of GM on patient outcomes. Cancer-related tests predominated (tumor profiling n = 9; germ-line testing n = 4), whereas the other eight reviews were divided among five topics. Twelve reviews discussed the importance of patient-centered outcomes, although most noted the limited direct evidence about these outcomes. Two reviews focused on delivery of genomic information to patients (communication of risk information and approaches to risk assessment). Most reviews (81%) explicitly identified comparators, which included no testing, other genomic tests, nongenomic tests, clinical criteria, and phenotype-based risk scores.
Cancer: tumor profiling for cancer diagnosis, prognosis, and/or treatment
Clinical context. Tumor profiling means testing tumor tissue for mutations or abnormal expression of gene products (GEP) that may be driving malignant behavior. Tumor profiling may classify the patient’s probability of recurrent disease or identify treatments that target the molecular mechanism of malignant growth.
Review topics. Review topics include urine-based tests for bladder and prostate cancer; gene expression tests for prostate, breast, and colon cancer; tests for cancers of unknown primary sites; prognostic tests for common cancers; genetic tests for cancer drug metabolism variants; and molecular tests to target cancer treatment.
Review conclusions ( N = 9) 19, 20, 21, 22, 23, 24, 25, 26, 27
-
• The best evidence for a gene expression profiling (GEP) test shows that OncotypeDx, which estimates the recurrence rate after surgery for early stage breast cancer, improves predictions based on clinical prognostic factors alone. Low to moderate risk of bias studies show that lower-risk OncotypeDX results are associated with lower rates of adjuvant chemotherapy.
-
• No published trials have prospectively measured the effect of OncotypeDX testing on clinical outcomes of early-stage breast cancer (clinical utility). A randomized trial (TAILORx)28 of adjuvant chemotherapy versus no chemotherapy is underway involving patients at intermediate risk for recurrence by OncotypeDX testing.
-
• In a 2013 review,20 tests for individual mutations (KRAS, ALK, EGFR, BRAF) and expression of multiple mRNA biomarkers (OncotypeDX, MammaPrint) improved prognostication when compared to clinical predictors (clinical validity). No study directly assessed whether these tests change downstream health outcomes, although OncotypeDX results did affect treatment decision making.
-
• A 2013 review25 of multigene panels to detect targeted therapy opportunities in advanced cancer found three prospective-retrospective studies using archival tumor samples. One compared outcomes of therapy matched to a single panel biomarker with unmatched therapy, whereas two had no controls.
-
• Studies should measure whether genomic tests lead to better clinical outcomes than alternative prognostic methods during different stages of common cancers.
Landscape analysis. The evidence base for using genomic tests to individualize cancer care is small. Good-quality evidence shows an effect on treatment choice in one test–cancer combination (OncotypeDX for recurrence of early-stage breast cancer). Furthermore, the MINDACT29 and TAILORx28 studies showed that the Mammaprint and OncotypeDX GEP panels predicted which early-stage breast cancer patients with low risk scores may be managed with endocrine therapy and avoid chemotherapy.29 The outcomes of these large trials involving breast cancer have been overall survival, disease-free survival, and survival without distant metastases. However, the RxPONDER trial (evaluation of OncotypeDX in node-positive patients) did involve patient advocates in the design of the study and includes quality of life and other patient-reported outcomes.11
Although GEP tests can estimate prognosis (clinical validity), CER should assess its effect on clinical outcomes (clinical utility). CER could also focus on colorectal, lung, skin, and hematologic cancers, for which tests have been developed and could be useful.30 The one review that assessed multiple molecular markers to target treatment found no high-quality evidence and highlighted the methodological complexities of designing unbiased studies to measure clinical utility.25
Cancer risk assessment
Clinical context. Detecting genes that are associated with an increased probability of developing cancer or harboring an undiagnosed cancer may lead to more intense surveillance or early treatment.
Review topics. Review topics include fecal DNA testing for colorectal cancer risk and genomic risk assessment for breast cancer risk.
Review conclusions ( N = 4) 31, 32, 33, 34
-
• Fecal DNA testing can detect colorectal cancer and large adenomas that are likely to become malignant. The only study of a currently marketed test measured sensitivity and specificity (clinical validity), but not the added impact of testing on clinical outcomes.34
-
• Short-term patient outcomes such as reduced distress and accuracy of perceived risk improve after breast cancer risk assessment.
-
• An intact chain of evidence leads from a strong family history of breast cancer to GM testing for BRCA1/2 to better outcomes after prophylactic bilateral mastectomy for BRCA mutation carriers,35,36 but no studies have directly measured the impact of BRCA testing on health outcomes. Existing studies lack real-world settings or diverse at-risk populations.
-
• Studies should examine consequences of testing for individuals and families, including acceptability to patients, adherence to screening, delivery of genomic testing, and models to estimate the incremental net benefit of testing and optimal testing intervals.
Landscape analysis. The list of inherited genetically determined variations in cancer risk is growing, as is the demand for genomic approaches to risk assessment. The included systematic reviews found no studies of clinical impact on disease outcomes. Targets for CER include family history and BRCA testing to identify breast/ovarian cancer risk and family histories of colorectal cancer and testing for mutations in the five genes associated with Lynch syndrome.30 The availability of several genomic tests for the same cancer presents opportunities for head-to-head comparisons. In addition to clinical outcomes, these studies should evaluate the acceptability of the tests and follow-up rates after abnormal test results.
Chronic conditions including neurodevelopmental delays
Clinical context. Among children with delayed cognitive development, identifying a gene that is associated with a specific condition has potential benefits, including informed reproductive decision making, a firmer prognosis, access to needed services, avoidance of unnecessary testing, and, possibly, improved health outcomes.
Review topics. Review topics include testing for developmental delay, intellectual disability, and autism spectrum disorder.
Review conclusions ( N = 2) 37, 38
-
• Observational, noncomparative studies have measured the yield of chromosomal microarray testing for gene copy number variants (which are more common in developmentally disabled children) and the actions taken by families. The effects of these actions and whether they would have occurred without GM testing are unknown.
-
• No study has compared the clinical outcomes of genomic testing for neurodevelopmental disorders with no testing.
-
• Comparative studies should compare positive and negative outcomes important to patients and their families, including the impact on reproductive decision making.
Landscape analysis. Chronic conditions have a high health burden and impact family members. Testing could potentially be useful for a large number of chronic conditions, but our selection of reviews addressed only neurodevelopmental delays in children (two reviews). Tests for familial hypercholesterolemia30 offer other opportunities for CER studies of the incremental benefits and harms of genomic testing. Because genes related to chronic diseases typically have low penetrance and high heterogeneity, assessing the impact of genomic testing on health outcomes will most likely require very large populations, suggesting the need for disease registries. Although the protracted course of chronic disease implies the need for long follow-up, the effect of genomic testing could lead relatively quickly to interventions that affect short-term patient-centered outcomes.
Pharmacogenetic testing
Clinical context. The metabolism of some drugs (conversion to an active form or an inactive form) is under genetic control. Mutations in the genes for enzymes that metabolize drugs may lead to too much or too little active drug when physicians prescribe standard doses. A pharmacogenetic test aims to detect a genetic basis for differences in the response to a drug.
Review topics. Review topics include testing for CYP2C19 genetic variants to guide antiplatelet therapy in coronary artery disease (clopidogrel) and testing for CYP2D6 genetic variants to guide tamoxifen therapy for women at high risk for primary breast cancer or recurrence.
Review conclusions ( N = 2) 39, 40
-
• A heterogeneous body of evidence regarding the effects of testing for CYP2C19 variants on clinical outcomes is insufficient to show that testing or CYP2C19 status alters cardiovascular event rates. Studies were small, had short-term outcomes, and seldom reported clinical outcomes.
-
• Trials of the impact of CYP2D6 testing on breast cancer outcomes have not been performed, probably because the evidence that variants in the CYP2D6 gene affect clinical outcomes is observational, inconsistent, and of only moderate quality.
-
• Studies should focus on standardizing testing methods and directly comparing the impact of testing strategies on patient-relevant clinical outcomes in large trials.
Landscape analysis. The two reviews identified only one small trial that measured the impact of pharmacogenetic testing on clinical outcomes. Since the completion of the systematic review, the National Heart, Lung, and Blood Institute and Mayo Clinic have been sponsoring a large pragmatic trial (TAILOR-PCI) to evaluate use of CYP2C19 genotyping to guide the choice between clopidogrel and ticagrelor. The purpose of this study was to determine whether using pharmacogenetic (PGx) testing to guide anti-platelet therapy improves cardiovascular outcomes following coronary stent placement in patients with impaired activation of clopidogrel.41 There are many other opportunities to conduct CER studies of PGx tests because nearly 200 drugs contain pharmacogenetic information.42 For example, three high-quality trials published in 2013 showed that, overall, a clinical algorithm was a better approach than PGx testing for dosing either warfarin or acenocoumarol and phenprocoumon and achieving desired states of anticoagulation.43,44,45 PGx testing might impact several conditions (e.g., infectious disease, mental health conditions, Stevens-Johnson syndrome).46 Further opportunities for CER include comparing PGx testing before starting treatment versus prescribing without testing. This strategy involves having access to PGx data at the point of care and requires access to clinical decision support but overcomes many of the logistical hurdles that have limited clinical integration of PGx testing.47
Prenatal screening
Clinical context. Aneuploidies (an abnormal number of autosomal or sex chromosomes) often lead to developmental abnormalities, the best known of which is Down syndrome (trisomy 21). Until recently, noninvasive tests for aneuploidies were not specific enough to act upon and therefore required tissue sampling for confirmatory karyotyping. Sequencing DNA from maternal serum can detect fragments of fetal DNA, which are present as early as 8–10 weeks of gestation.
Review topics. Review topics include sequencing-based tests for fetal Down syndrome (trisomy 21) and other aneuploidies.
Review conclusions ( N = 2) 48, 49
-
• The goals of the studies were to measure sensitivity, measure specificity, and measure posttest probabilities, but not to measure the clinical impact of fetal DNA testing versus other noninvasive tests for aneuploidies.
-
• Tests for aneuploidies in maternal serum are nearly 100% specific (rare false-positive results) and more sensitive than conventional tests. The study’s risk of bias was low.
-
• The posttest probabilities of aneuploidies after positive and negative test results were better than conventional screening tests, thereby providing indirect evidence of better clinical utility for DNA sequencing–based tests for trisomy 21, trisomy 18, and trisomy 13.
-
• No studies measured patient-centered outcomes.
-
• Future studies should compare all potential screening strategies and prospectively examine outcomes in average-risk populations and screening for other chromosomal abnormalities.
Landscape analysis. Prenatal screening for fetal chromosomal abnormalities is considered the standard of care in the United States for women at high risk. Because of its very high specificity, cell-free DNA screening is a strong alternative to testing for maternal serum markers and fetal ultrasound and should result in fewer invasive procedures to perform karyotyping. CER studies could compare parent preferences for further testing after learning the results of DNA testing versus conventional noninvasive testing, explore ethical questions, and study patient, family, and system effects of expanding prenatal testing to average-risk populations.
Population screening for risk assessment
Clinical context. DNA-based tests can detect genes that increase the risk of diseases like breast cancer, lung cancer, or diabetes. In some cases, detecting such genes could lead to preventive measures (e.g., bilateral mastectomy for carriers of BRCA mutations, which increases the risk of breast cancer) or changes in risky behaviors.
Review topics. Review topics include effects of communicating DNA-based risk estimates on risk-reducing behaviors.
Review conclusions ( N = 1) 50
-
• Larger and better-quality randomized, control trials should examine patient-centered outcomes, including behavior and unintended adverse effects. Sample sizes should be large enough to detect small effects.
-
• Poor-quality evidence suggests that communicating genetic disease risk has little or no impact on smoking or physical exercise.
-
• No studies had a low risk of bias or compared genomic test and conventional risk factor counseling versus counseling alone.
Landscape analysis. Although risky behaviors are a major health threat, current evidence does not support claims that DNA-based risk assessments motivate behavior change. The key unanswered question is the added effect of genomic risk information on clinical outcomes when combined with and compared to conventional testing, family history, clinical risk factors, and treatment. Such studies require a comparison group that receives conventional care but no genomic testing. Several randomized trials involved in the only systematic review that focused on provision of genetic information to reduce risk behaviors (e.g., smoking, diet, physical activity) did meet this study design criterion but were of low quality. The best approach to assessing the impact of genomic testing on risky behaviors such as cigarette smoking may be to study screening for mutations in highly penetrant genes in patients with a target condition for which effective interventions exist. Such studies should examine whether genomic information has an incremental impact on behavior and outcomes by using carefully designed comparators.
Whole-exome sequencing and whole-genome sequencing and testing for rare diseases
Clinical context. Disorders that present with multiple anomalies, often early in life, suggest a mutation in a single gene. These disorders can be difficult to diagnose because of their rarity, nonspecific phenotype, and lack of a well-defined pathway to a diagnosis. Establishing a genetic cause can lead to specific treatment and to establishing the carrier status of family members.
Review topics. Review topics include sequencing for disorders caused by a single gene.
Review conclusions ( N = 1) 51
-
• Sequencing may end “diagnostic odysseys” for patients and inform reproductive decisions, but the only information regarding clinical utility is anecdotal (no systematic study of clinical impact).
-
• No studies of the broader uses of sequencing tests.
-
• Ethical issues, such as the consequences of pursuing secondary findings, require study.
Landscape analysis. Opportunities for CER studies may increase as the indications for whole-exome sequencing (WES) and whole-genome sequencing (WGS) expands. Testing only to diagnose a suspected rare disease will have a small public health impact due to low prevalence and lack of treatments. Evidence of clinical utility is lacking for the broader use of WES and WGS beyond diagnosis of rare diseases. The American College of Medical Genetics and Genomics guidelines on the return of secondary findings from WES and WGS list some potentially important tests for CER studies such as detection of genetic familial hypercholesterolemia.52,53
Discussion
In summary, we found a very limited body of evidence about the effect of using genomic tests on health outcomes and many evidence gaps for CER to address. Like the systematic reviews, we defined clinical utility as improved health outcomes triggered by a test result and mediated by changes in patient behavior and clinical decisions. We also found a lack of evidence for effects on intermediate outcomes (e.g., avoiding unnecessary care, improving access to services, or providing prognostic or predictive information), which negates efforts to build a chain of evidence from test results through these intermediate outcomes to altered clinical outcomes.
Key implications of our findings for research and policy are presented here.
Address important questions
Two important decisions relevant to CER involve whether to perform a test and what to do with patients at intermediate risk or with uncertain test results. The first decision addresses the question “Is the test result likely to change the treatment plan suggested by my clinical assessment?” A CER study design might compare health-related outcomes after the clinical assessment alone with outcomes after performing the assessment plus a GM test. The second decision is in regard to what to do in ambiguous situations. A study design might randomly assign patients with intermediate-risk test results to receive either low-intensity treatment or high-intensity treatment.
Diversify the evidence base
Most of the reviews focused on cancer; however, many oncology-specific GM tests known to have promising evidence supporting their use in clinical practice were not included in the reviews because of either their scope or their timing. In addition to oncology, active areas of research in neurology, psychiatry, cardiology, and rare disorders suggest opportunities for future studies and evidence syntheses.54 The Centers for Disease Control and Prevention’s Office of Public Health Genomics categorize genetic tests in three tiers according to whether they have a base of synthesized evidence.30 The agency’s list of tests categorized as “Tier 2” would be attractive targets for future CER studies because they are mentioned in clinical practice guidelines or Food and Drug Administration labeling, but the supporting evidence is insufficient to guide clinical use. Finally, even the best studies focused on how test results affected decision-making by clinicians rather than by patients. Missing are high-quality studies comparing the effect of different models for communicating DNA-based disease risk estimates on patient motivation to take appropriate actions.
Use established methods to improve the evidence base
We found only a few randomized trials designed to assess the impact of GM testing on patient outcomes. However, observational studies (cohort, retrospective-prospective, single arm) and use of indirect evidence (modeling) can also compare clinical utility, albeit with less certain results. Studies to establish analytic and clinical validity should precede studies to measure clinical utility to ensure that study participants are exposed to accurate and reliable tests that could change patient care. Many of the included tests had good evidence regarding analytical validity but lacked a chain of evidence leading from test results to clinical outcomes. Several publications have provided methodological guidance for studying genomic tests.55,56,57 GM has some characteristics that may require more complex analyses than other types of interventions (e.g., analyzing the impact of inherited mutations on family members), although conventional study designs can still be used. Using established methods could reduce concerns that GM is subject to higher evidence standards than other interventions.
Include outcomes that matter to patients
For genomic tests to become routine practice, using them should favorably impact outcomes that matter to patients. To determine whether GM is fulfilling its promise, patient-centered CER should compare GM tests plus non-GM risk assessment versus non-GM risk assessment alone, other GM tests, or both. Research team leaders should use real-world settings and seek advice from patients about which outcomes matter most.58
Use consistent and unbiased study methods
In the future, the evidence for individualized patient care will increasingly come from statistically reliable subgroup analyses derived from patient-level meta-analyses. These meta-analyses require many large, high-quality studies. Some evidence gaps in the systematic reviews were due to poor-quality research that could not support any conclusions. Moreover, the studies used diverse study designs, tests, study populations, and outcome measures, which would make it difficult to draw conclusions from a pooled study population. To raise the standard of evidence, GM researchers should cooperate to establish study quality standards.
Keep pace with technology changes
As GM evolves beyond single-gene tests, CER studies must keep pace. Only two of our systematic reviews covered multigene tests, WES, and WGS. Multigene tests are complex and evolving. A recent National Academy of Medicine report noted the obstacles and recommended several approaches to address them.59 Tumor-agnostic, biomarker-driven clinical trials such as NCI-MATCH, SWOG’s LUNG-MAP, and the American Society of Clinical Oncology’s TAPUR are evidence of progress. Recent publications about WES in rare diseases, cancer, and complex disorders suggest the need to address current evidence gaps about personal and clinical utility soon.53,60
Our review suggests opportunities to close important evidence gaps about using genomic tests; however, additional methods-development work is needed. Methodological topics that need ongoing stakeholder-informed dialogue include the following.
Patient preferences
The purpose of CER is to design and conduct studies that meet the information needs of patients, clinicians, and policymakers facing decisions. Engaging these stakeholders as partners in all phases of the research process should ensure that future CER meets their information needs. Meaningful engagement of patients in GM CER will require attention to numeracy and genetic literacy and the willingness to engage in shared decision-making about performing genomic tests and their interpretation. For example, noninvasive prenatal screening tests for fetal chromosomal abnormalities (i.e., trisomy 21, 18, and 13) have become standard clinical practice on the basis of their clinical validity and private insurer coverage.48 However, questions remain about whether clinicians and patients fully understand the risks and benefits of using these tests, which also detect sex chromosome aneuploidies and microdeletions, as compared to alternative screening methods that do not detect them. Also, we lack research about how patients and clinicians make decisions after positive test results and about between-partner concordance about pregnancy termination decisions in patient subgroups stratified by maternal risk, race, ethnicity, or socioeconomic status.61,62 Another issue is that outcomes of WES and WGS may lead to gains in well-being that go beyond the impact on morbidity and mortality (e.g., reassurance or providing a diagnosis for an untreatable condition).51 GM tests may also harm. CER questions to address might include whether WES tests increase personal well-being beyond conventional testing and genetic counseling and under what circumstances. The answers to these questions might inform the debate about whether to consider personal utility when developing practice guidelines and coverage policies.
Behavior change strategies
Changing risky behaviors is difficult, and the motivation to change is part of the problem.50 Of particular relevance to common, chronic conditions is whether current risk assessment approaches such as obtaining family histories are sufficient or whether taking the next step and performing genetic testing lead to better health outcomes.38 In the field of cardiovascular disease, CER questions involve the following: the net benefits to patients, families, and society from adding genetic information on cardiovascular disease to conventional risk assessment approaches, such as family risk history and clinical risk scores, and whether providing genetic information in addition to information about smoking, hypertension, and hypercholesterolemia leads to adopting a healthier lifestyle or more comprehensive preventive therapy and, ultimately, better cardiac outcomes. Trials to date have shown little or no effect on health-risk behaviors.
Value
With decreasing costs of next-generation sequencing, proponents of multigene panels have argued that it is more efficient to test for multiple mutations simultaneously rather than to conduct multiple single-gene tests. High-quality evidence is lacking.63 Advocates for using multigene panels for tumor profiling argue that future treatment advances depend on an understanding of tumor biology gained through tumor biomarker panels, not conventional classifications based on tumor histology, grade, and stage.64 Key questions include the optimal gene panel size, how to select treatments based on presumed driver mutations, and the role of tumor genome heterogeneity.25,65,66 CER could address the key question regarding how often patients are matched to potential treatments using large, tumor-agnostic multigene panels and the clinical outcomes versus tumor-specific single-gene testing and clinical factors alone.
Our study had limitations. We examined systematic reviews rather than individual CER studies, and we selected systematic reviews commissioned or conducted by TAGs. Thus, our results do not represent the entire universe of CER studies of GM. This strategy did mean that experienced teams of systematic reviewers performed the included reviews using standardized definitions of study quality, which was important to one of our principal goals: to form credible judgment regarding the quality of CER for GM. Although the TWG found the conclusions of the reviews to be representative of the broader literature, we did not systematically search for recent articles on these topics. Although we included a range of TAGs in our search, 2 of them (AHRQ and BCBSA TEC) produced all but 2 of the 21 included systematic reviews and therefore provide a predominately United States–focused perspective. Finally, to address our study objectives, we had to categorize and summarize across disparate reviews and augment the review findings with expert opinions. Therefore, readers should not consider our results definitive. Still, the included systematic reviews cover most of the spectrum of clinical topics addressed by GM. Future research should examine the cost-effectiveness and budget impact and how evidence can be aligned with the current use of tests in the best way possible.
In conclusion, our findings can inform decisions about where to focus future research and policy initiatives. Over the next few decades, patients, clinicians, and policymakers will be asking whether the added information provided by GM leads to better health outcomes than using conventional clinical information, such as family history and non-GM tests. CER is the research design for answering these questions.
Disclosure
This study was funded in part by a National Human Genome Research Institute grant (R01HG007063 to K.A.P.) and a National Cancer Institute grant to the UCSF Helen Diller Family Comprehensive Cancer Center (5P30CA082013). In addition, research reported in this publication was partially funded through PCORI contracts (to K.A.P., P.A.D., S.R.T., and L.A.O.). H.C.S. is an employee of PCORI. The other authors declare no conflict of interest.
References
Ginsburg GS, Kuderer NM. Comparative effectiveness research, genomics-enabled personalized medicine, and rapid learning health care: a common bond. J Clin Oncol 2012;30:4233–4242.
Khoury MJ, Rich EC, Randhawa G, Teutsch SM, Niederhuber J. Comparative effectiveness research and genomic medicine: an evolving partnership for 21st century medicine. Genet Med 2009;11:707–711.
Deverka PA, Haga SB. Comparative effectiveness research and demonstrating clinical utility for molecular diagnostic tests. Clin Chem 2015;61:142–144.
IJzerman M, Manca A, Keizer J, Ramsey S. Implementation of comparative effectiveness research in personalized medicine applications in oncology: current and future perspectives. J Comp Eff Res. 2015;5:65–72.
Ramsey SD, Veenstra D, Tunis SR, Garrison L, Crowley JJ, Baker LH. How comparative effectiveness research can help advance ‘personalized medicine’ in cancer treatment. Health Aff (Millwood) 2011;30:2259–2268.
Thariani R, Wong W, Carlson JJ, et al.; Center for Comparative Effectiveness Research in Cancer Genomics (CANCERGEN). Prioritization in comparative effectiveness research: the CANCERGEN Experience. Med Care 2012;50:388–393.
Phillips KA, Trosman JR, Kelley RK, Pletcher MJ, Douglas MP, Weldon CB. Genomic sequencing: assessing the health care system, policy, and big-data implications. Health Aff (Millwood) 2014;33:1246–1253.
Phillips KA. Closing the evidence gap in the use of emerging testing technologies in clinical practice. JAMA 2008;300:2542–2544.
Institute of Medicine of the National Academies. Committee on Standards for Systematic Reviews of Comparative Effectiveness Research. Eden J, Levit L, Berg A, Morton S (eds). Finding What Works in Health Care: Standards for Systematic Reviews. National Academies Press: Washington, DC, 2011.
Thariani R, Wong W, Carlson JJ, et al.; Center for Comparative Effectiveness Research in Cancer Genomics (CANCERGEN). Prioritization in comparative effectiveness research: the CANCERGEN Experience. Med Care 2012;50:388–393.
Ramsey SD, Barlow WE, Gonzalez-Angulo AM, et al. Integrating comparative effectiveness design elements and endpoints into a phase III, randomized clinical trial (SWOG S1007) evaluating oncotypeDX-guided management for women with breast cancer involving lymph nodes. Contemp Clin Trials 2013;34:1–9.
Agency for Healthcare Research and Quality: Archived EPC Evidence Reports. http://archive.ahrq.gov/clinic/epcarch.htm. Accessed 9 September 2016.
PCORI: Generation and Prioritization of Topics for Funding Announcements. 2015. http://www.pcori.org/research-results/how-we-select-research-topics/generation-and-prioritization-topics-funding-4. Accessed 13 April 2016.
Gil MM, Quezada MS, Revello R, Akolekar R, Nicolaides KH. Analysis of cell-free DNA in maternal blood in screening for fetal aneuploidies: updated meta-analysis. Ultrasound Obstet Gynecol 2015;45:249–266.
Gregg AR, Skotko BG, Benkendorf JL, et al. Noninvasive prenatal screening for fetal aneuploidy, 2016 update: a position statement of the American College of Medical Genetics and Genomics. Genet Med 2016;18:1056–1065.
Langlois S, Brock JA, Wilson RD, et al.; Genetics Committee. Current status in non-invasive prenatal detection of Down syndrome, trisomy 18, and trisomy 13 using cell-free DNA in maternal plasma. J Obstet Gynaecol Can 2013;35:177–183.
Norton ME, Wapner RJ. Cell-free DNA analysis for noninvasive examination of trisomy. N Engl J Med 2015;373:2582.
Zhang H, Gao Y, Jiang F, et al. Non-invasive prenatal testing for trisomies 21, 18 and 13: clinical experience from 146,958 pregnancies. Ultrasound Obstet Gynecol 2015;45:530–538.
Chou R, Buckley D, Fu R, et al. Emerging Approaches to Diagnosis and Treatment of Non–Muscle-Invasive Bladder Cancer. AHRQ: Pacific Northwest Evidence-based Practice Center. 25 October 2015.
Meleth S, Reeder-Hayes K, Ashok M, et al. Technology Assessment of Molecular Pathology Testing for the Estimation of Prognosis for Common Cancers. AHRQ: RTI International–University of North Carolina at Chapel Hill Evidence-based Practice Center. 29 May 2014.
Bradley L, Palomaki G, Gutman S, Samson D, Aronson N. PCA3 Testing for the Diagnosis and Management of Prostate Cancer. AHRQ: Blue Cross and Blue Shield Association Technology Evaluation Center Evidence-based Practice Center. April 2013.
Meleth S, Whitehead N, Swinson-Evans T, Lux L. Technology Assessment on Genetic Testing or Molecular Pathology Testing of Cancer with Unknown Primary Site to Determine Origin. AHRQ: RTI International-University of North Carolina at Chapel Hill Evidence-based Practice Center. 20 February 2013.
Ratko T. Gene Expression Analysis for Prostate Cancer Management. Blue Cross and Blue Shield Technology Evaluation Center. January 2015.
Glacy J, Chopra R. Gene Expression Profiling in Women with Lymph Node-Negative Breast Cancer to Select Adjuvant Chemotherapy. Blue Cross and Blue Shield Technology Evaluation Center. October 2014.
Mark D, Ratko T. Special Report: Multiple Molecular Testing of Cancer to Identify Targeted Therapies. Blue Cross and Blue Shield Technology Evaluation Center. June 2013.
Black E, Falzon L, Aronson N. Gene Expression Profiling for Predicting Outcomes in Stage II Colon Cancer. AHRQ: Blue Cross and Blue Shield Association Technology Evaluation Center Evidence-based Practice Center. December 2012.
Terasawa T, Dahabreh I, Castaldi P, Trikalinos T. Systematic Reviews on Selected Pharmacogenetic Tests for Cancer Treatment: CYP2D6 for Tamoxifen in Breast Cancer, KRAS for anti-EGFR antibodies in Colorectal Cancer, and BCR-ABL1 for Tyrosine Kinase Inhibitors in Chronic Myeloid Leukemia. AHRQ: Tufts Evidence-based Practice Center. 7 June 2010.
Sparano JA, Gray RJ, Makower DF, et al. Prospective validation of a 21-gene expression assay in breast cancer. N Engl J Med 2015;373:2005–2014.
Cardoso F, van’t Veer LJ, Bogaerts J, et al.; MINDACT Investigators. 70-gene signature as an aid to treatment decisions in early-stage breast cancer. N Engl J Med 2016;375:717–729.
Centers for Disease Control and Prevention. Genetic Testing—Genomic Tests and Family Health History by Levels of Evidence. 2015. http://www.cdc.gov/genomics/gtesting/tier.htm. Accessed 12 April 2016.
Nelson H, Fu R, Goddard K, et al. Risk Assessment, Genetic Counseling, and Genetic Testing for BRCA-related Cancer: Systematic Review to Update the U.S. Preventive Services Task Force Recommendation. AHRQ: Pacific Northwest Evidence-based Practice Center. December 2013.
Lin J, Webber E, Beil T, Goddard K, Whitlock E. Fecal DNA Testing in Screening for Colorectal Cancer in Average-Risk Adults. AHRQ: Oregon Evidence-based Practice Center. 29 February 2012.
Mark D. Fecal DNA Analysis for Colorectal Cancer Screening. Blue Cross and Blue Shield Technology Evaluation Center. December 2014.
Hilgart J, Coles B, Iredale R. Cancer genetic risk assessment for individuals at risk of familial breast cancer. Cochrane Database Syst Rev 2012;CD003721.
Rebbeck TR, Friebel T, Lynch HT, et al. Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group. J Clin Oncol 2004;22:1055–1062.
Moyer VA ; US Preventive Services Task Force. Risk assessment, genetic counseling, and genetic testing for BRCA-related cancer in women: U.S. Preventive Services Task Force recommendation statement. Ann Intern Med 2014;160:271–281.
Grant M, Chopra R. Special Report: Chromosomal Microarray for the Genetic Evaluation of Patients With Global Developmental Delay, Intellectual Disability, and Autism Spectrum Disorder. Blue Cross and Blue Shield Technology Evaluation Center. August 2015.
Sun F, Oristaglio J, Levy S, et al. Genetic Testing for Developmental Disabilities, Intellectual Disability and Autism Spectrum Disorder. AHRQ: ECRI Institute–Penn Medicine Evidence-based Practice Center. 29 June 2015.
Dahabreh I, Moorthy D, Lamont J, Chen M, Kent D, Lau J. Testing of CYP2C19 Variants and Platelet Reactivity for Guiding Antiplatelet Treatment. AHRQ: Tufts Evidence-based Practice Center. 25 September 2013.
Glacy J. CYP2D6 Pharmacogenomics of Tamoxifen Treatment. Blue Cross and Blue Shield Technology Evaluation Center. January 2014.
Klein T. Mayo Clinic Studying Genomics of Antiplatelet Heart Medication. 2016. http://newsnetwork.mayoclinic.org/discussion/mayo-clinic-studying-genomics-of-antiplatelet-heart-medication-2644cc/.
PharmGkb. Drug Labels. 2016. https://www.pharmgkb.org/view/drug-labels.do. Accessed 2 November 2016.
Kimmel SE, French B, Kasner SE, et al.; COAG Investigators. A pharmacogenetic versus a clinical algorithm for warfarin dosing. N Engl J Med 2013;369:2283–2293.
Pirmohamed M, Burnside G, Eriksson N, et al.; EU-PACT Group. A randomized trial of genotype-guided dosing of warfarin. N Engl J Med 2013;369:2294–2303.
Verhoef TI, Ragia G, de Boer A, et al.; EU-PACT Group. A randomized trial of genotype-guided dosing of acenocoumarol and phenprocoumon. N Engl J Med 2013;369:2304–2312.
Plumpton CO, Roberts D, Pirmohamed M, Hughes DA. A systematic review of economic evaluations of pharmacogenetic testing for prevention of adverse drug reactions. Pharmacoeconomics 2016;34:771–793.
Bielinski SJ, Olson JE, Pathak J, et al. Preemptive genotyping for personalized medicine: design of the right drug, right dose, right time-using genomic data to individualize treatment protocol. Mayo Clin Proc 2014;89:25–33.
Ratko T, Chopra R. Noninvasive Prenatal Cell-Free Fetal DNA-based screening for Aneuploidies Other Than Trisomy 21. Blue Cross and Blue Shield Technology Evaluation Center. December 2014.
Piper M, Civic D, Grant M, Lefevre F. Sequencing-Based Tests to Determine Fetal Down Syndrome (Trisomy 21) form Maternal Plasma DNA. Blue Cross and Blue Shield Technology Evaluation Center. April 2013.
Marteau T, French D, Griffin S, et al. Effects of communicating DNA-based disease risk estimates on risk-reducing behaviors . Cochrane Database Syst Rev 2010;CD007275.
Piper M, Grant M. Special Report: Exome Sequencing for Clinical Diagnosis for Patients with Suspected Genetic Disorders. Blue Cross and Blue Shield Technology Evaluation Center. August 2013.
Green RC, Berg JS, Grody WW, et al.; American College of Medical Genetics and Genomics. ACMG recommendations for reporting of incidental findings in clinical exome and genome sequencing. Genet Med 2013;15:565–574.
ACMG Board of Directors. ACMG policy statement: updated recommendations regarding analysis and reporting of secondary findings in clinical genome-scale sequencing. Genet Med 2015;17:68–69.
Schully SD, Lam TK, Dotson WD, et al. Evidence synthesis and guideline development in genomic medicine: current status and future prospects. Genet Med 2015;17:63–67.
Institute of Medicine of the National Academies. Micheel CM, Nass SJ, Omenn GS (eds). Evolution of Translational Omics: Lessons Learned and the Path Forward. National Academies Press: Washington, DC, 2012.
Deverka P, Messner DA, McCormack R, et al. Generating and evaluating evidence of the clinical utility of molecular diagnostic tests in oncology. Genet Med 2016;18:780–787.
Parkinson DR, McCormack RT, Keating SM, et al. Evidence of clinical utility: an unmet need in molecular diagnostics for patients with cancer. Clin Cancer Res 2014;20:1428–1444.
Basch E, Spertus J, Dudley RA, et al. Methods for Developing Patient-Reported Outcome-Based Performance Measures (PRO-PMs). Value Health 2015;18:493–504.
National Academies of Sciences, Engineering, and Medicine. Biomarker Tests for Molecularly Targeted Therapies: Key to Unlocking Precision Medicine. National Academies Press: Washington, DC, 2016.
Tetreault M, Bareke E, Nadaf J, Alirezaie N, Majewski J. Whole-exome sequencing as a diagnostic tool: current challenges and future opportunities. Expert Rev Mol Diagn 2015;15:749–760.
Weitzel KW, Elsey AR, Langaee TY, et al. Clinical pharmacogenetics implementation: approaches, successes, and challenges. Am J Med Genet C Semin Med Genet 2014;166C:56–67.
Chang KL, Weitzel K, Schmidt S. Pharmacogenetics: using genetic information to guide drug therapy. Am Fam Physician 2015;92:588–594.
Saffitz JE. Genomic pathology: a disruptive innovation. Medicine (Baltimore). 2012:237–239.
Meric-Bernstam F, Farhangfar C, Mendelsohn J, Mills GB. Building a personalized medicine infrastructure at a major cancer center. J Clin Oncol 2013;31:1849–1857.
Trosman JR, Weldon CB, Kelley RK, Phillips KA. Challenges of coverage policy development for next-generation tumor sequencing panels: experts and payers weigh in. J Natl Compr Canc Netw 2015;13:311–318.
Phillips K, Trosman J, Weldon C, Chambers J, Deverka P, Douglas M. Few insurer policies currently cover multi-gene panels: findings from a systematic registry. Nat Biotechnol, in press.
Acknowledgements
We gratefully acknowledge the experts who provided background information and participated in expert interviews: James P. Evans, Robert Nussbaum (Invitae and UCSF), Sharon F. Terry, (Genetic Alliance), and Bruce Quinn (Bruce Quinn Associates). We also acknowledge insightful comments from Stanley Ip (PCORI). None of the consulting experts received compensation, and each provided written permission to be to acknowledged. One author (H.S.) is an employee of the Patient-Centered Outcomes Research Institute (PCORI). All statements in this report, including its findings and conclusions, are solely the responsibility of the authors and do not necessarily represent the views of PCORI, its Board of Governors, or its Methodology Committee. Nothing in this manuscript should be inferred to represent the viewpoints of the National Institutes of Health.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
(DOCX 113 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/4.0/
About this article
Cite this article
Phillips, K., Deverka, P., Sox, H. et al. Making genomic medicine evidence-based and patient-centered: a structured review and landscape analysis of comparative effectiveness research. Genet Med 19, 1081–1091 (2017). https://doi.org/10.1038/gim.2017.21
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/gim.2017.21
Keywords
This article is cited by
-
Evaluating genetic and genomic tests for heritable conditions in Australia: lessons learnt from health technology assessments
Journal of Community Genetics (2022)
-
Quality of life drives patients’ preferences for secondary findings from genomic sequencing
European Journal of Human Genetics (2020)
-
Clinical genomic testing: what matters to key stakeholders?
European Journal of Human Genetics (2020)
-
Rapid whole-genome sequencing decreases infant morbidity and cost of hospitalization
npj Genomic Medicine (2018)
-
A proposed approach to accelerate evidence generation for genomic-based technologies in the context of a learning health system
Genetics in Medicine (2018)